Introduction
The search for a red-emitting fluorescent protein with performance attributes similar to those of the enhanced green fluorescent protein (EGFP) from the Aequorea victoria jellyfish (in effect, brightness, photostability, and utility in fusions) has been seen as a critical avenue to providing an important tool for multicolor imaging and in generating new fluorescence resonance energy transfer (FRET) biosensors with spectral profiles in the longer wavelength regions. Another driver for the development of red fluorescent proteins is that cellular autofluorescent is significantly reduced in this spectral region, allowing the probes to be detected deeper into biological tissue. Furthermore, living cells and tissues better tolerate illumination by the longer excitation wavelengths, allowing extend periods for imaging. Unfortunately, after years of unsuccessful mutagenesis attempts to develop red fluorescent proteins (RFPs) from the Aequorea-based fluorescent proteins, the yellow fluorescent proteins (YFPs) remain the most red-shifted of the GFP derivatives.
To address this problem, several groups of investigators demonstrated that much of the color diversity in reef corals is a result of GFP-like proteins. It is widely thought that these proteins evolved in the corals to fulfill roles that are distinct from those in the luminescent jellyfish. For example, the production of fluorescent proteins and related chromoproteins (fluorescent protein-like proteins that absorb but do not fluoresce) in the sessile corals might function in a photo-protective role. Alternatively, the fluorescent proteins might function to support the symbiotic relationships between the corals and algae. Regardless of their evolutionary origin or biological function, many corals and other Anthozoa species have been identified that produce proteins that are very closely related to fluorescent proteins. These marine organisms are currently being mined for novel proteins in order to fill the gaps in fluorescent protein color palette and improve overall performance.
Over the past decade, fluorescent proteins spanning the entire visible light spectrum have been characterized and cloned from Anthozoa, and some of these have been optimized for imaging applications. One of the first Anthozoa-derived fluorescent proteins to be extensively characterized was isolated from the sea anemone Discosoma striata. This novel fluorescent protein was originally called drFP583, but is now commonly known as DsRed. When expressed in cells, the fully matured DsRed fluorescent protein is optimally excited at 558 nanometers, and has an emission maximum at 583 nanometers. However, there are multiple problems associated with DsRed when used for live cell imaging. The maturation of DsRed is slow, and proceeds through an intermediate chromophore stage where most of the fluorescence emission occurs in the green spectral region. This “green state” introduces signal crosstalk that limits the utility of DsRed for multiple labeling experiments. In addition, DsRed is an obligate tetramer (see Figure 2) with the tendency to form oligomers, which can lead to protein aggregation and interference with the localization of protein fusions in living cells. Although these side effects are not important when the probe is utilized simply as a reporter for gene expression, the application of DsRed for a wide variety of investigations in cell biology is severely compromised. Thus, in contrast to the Aequorea-based fluorescent proteins that have been used to successfully label hundreds of different targets, protein fusions to DsRed have proven far more problematic. These shortcomings have required strategies to improve DsRed through mutagenesis, or alternatively, to search for other fluorescent proteins from corals with more optimal characteristics for live cell imaging.
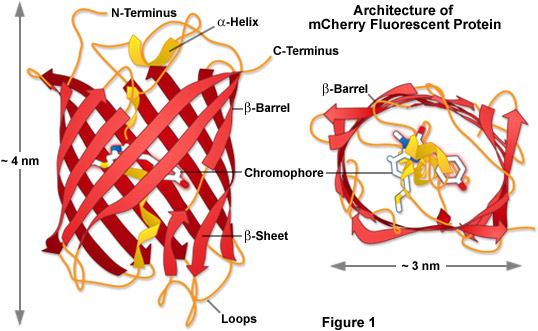
Presented in Figure 1 is the molecular architecture of a high-performance monomeric Anthozoa fluorescent protein named mCherry, which was derived from DsRed through extensive mutagenesis as described in detail below. These drawings were modeled using information available from the Protein Data Bank (identification number 2h5q). The beta-barrel structure of Anthozoa fluorescent proteins is very similar to those found in Aequorea GFP variants in terms of the general features and dimensions despite rather low correlations in the amino acid sequence homology. Unlike the jellyfish proteins, however, those obtained from Anthozoa feature a barrel that is more elliptical in geometry when viewed from the top or bottom (see Figure 1), perhaps arising from the fact that most wild-type fluorescent proteins in this class were originally tetrameric in character (Figure 2). In Figure 1, the mCherry chromophore is positioned near the central axis of the beta-barrel in a portion of the alpha-helix that threads its way through the center of the structure.
back to top ^Fluorescent Protein Variants Based on DsRed
Several of the major problems associated with DsRed were overcome by site-directed and random mutagenesis approaches. This effort yielded a second-generation version of DsRed, appropriately called DsRed2, which contains a series of silent nucleotide substitutions corresponding to human codon preferences, as well as several mutations that increase the maturation rate. In addition, the elimination of a string of basic amino acid residues at the amino terminus of DsRed2 (by mutation to acidic or neutral moieties) significantly reduces the tendency of the protein to form aggregates. DsRed2 still forms a tetramer in solution, but the increased maturation rate greatly reduces the level of the intermediate green species, making it is more useful for multiple labeling experiments. Further enhancement in the rate of maturation was realized with the third generation of DsRed mutants, which also display an increased intrinsic brightness. For example, the DsRed-Express variant (available from Clontech) can be detected within an hour after transfection of cells, compared to approximately six hours for DsRed2 and 11 to 15 hours for DsRed. More advanced versions of DsRed, named DsRed-Express2 and DsRed-Max, exhibit faster maturation rates and improved solubility. However, since these direct descendents of DsRed remain obligate tetramers, there has been a concerted effort to generate dimeric and monomeric red fluorescent protein variants.
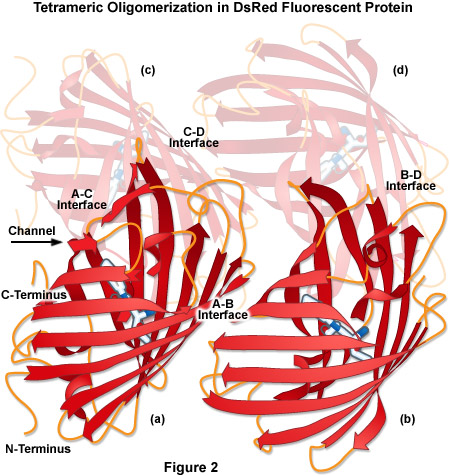
The tetrameric crystal structure of DsRed (Protein Data Bank identification number 1g7k) is illustrated in Figure 2. Despite having only 23 percent sequence homology with Aequorea victoria wild-type GFP, DsRed presents a tertiary fold that is very similar, containing an 11-stranded beta-barrel with a coaxial helix containing the chromophore. Among the major features of DsRed is the extremely close-packed tetrameric association of the four protomer beta-barrels that feature a small elliptical channel directly through the center of the tetramer. This region is lined with polar amino acid residues and salt bridges to accommodate water molecules that are bound between the individual units. The four monomeric units are arranged as a dimer of dimers where the A and C chains participate in close contacts via their surfaces and C-termini. In contrast, the A and B chains interact through large buried patches of hydrophobic residues with only a few salt bridges and hydrogen bonds.
The generation of truly monomeric DsRed variants, as well as monomers from proteins derived from a host of different Anthozoa species, has proven to be a difficult task. For example, site-directed mutagenesis to break the tetramer formation by DsRed2 resulted in the generation of a non-fluorescent monomer. To rescue fluorescence, investigators applied successive rounds of random mutagenesis to the monomer, selecting for proteins in each round with improved red fluorescence (this approach is called directed evolution). A total of 33 amino acid substitutions were required to generate the first-generation monomeric red fluorescent protein, which was termed mRFP1. The rapidly maturing mRFP1 overcame many critical problems associated with DsRed, while shifting the fluorescence emission about 25 nanometers deeper into the red spectrum. Unfortunately, mRFP1 exhibits significantly reduced fluorescence emission intensity (as expected, the quantum yield of the monomer is about 25 percent of DsRed2) and it is very sensitive to photobleaching. Furthermore, mRFP1 has an absorbance peak at 503 nanometers that arises from a non-fluorescent species, which indicates that a significant fraction of the protein never fully matures. Over the past several years, extensive mutagenesis efforts, including novel techniques such as iterative somatic hypermutation, have successfully been applied to mRFP1 to yield a new generation of orange, red, and far-red fluorescent protein variants.
back to top ^The mFruit Series of Fluorescent Proteins
One of the most productive developments in the crusade to generate useful fluorescent proteins in the orange and red spectral regions resulted from the directed evolution of mRFP1. Nathan Shaner and Roger Tsien speculated that the chromophore amino acids, Gln66 and Tyr67, which are critical determinants of the spectral characteristics of the Aequorea proteins, would play a similar role in determining the color of mRFP1 derivatives as well. In this case, the directed evolution approach was applied to derivatives of mRFP1, targeting these amino acid residues followed by selecting for new color variants. The result was a group of six new monomeric fluorescent proteins exhibiting emission maxima ranging from 540 nanometers to 610 nanometers. These new fluorescent proteins were named mHoneydew, mBanana, mOrange, mTangerine, mStrawberry, and mCherry, referencing the common fruits that bear colors similar to their respective emission profiles. Thus, these new fluorescent proteins are commonly known as the mFruits. Although the mFruits were a tour de force, yielding tremendous information about fluorescent protein structure and function, several mFruit fluorescent proteins, including mHoneydew, mBanana, and mTangerine, suffer from low intrinsic brightness and poor photostability.
The most promising members in the mFruit series are mOrange, mStrawberry, and mCherry. The mOrange variant is the brightest of the mFruit proteins and has spectral characteristics allowing it to be paired with other fluorescent proteins in the cyan and green spectral region for multicolor imaging and as a potential FRET acceptor. Unfortunately, the photostability of mOrange is only approximately 5 percent that of EGFP. Recently, this deficiency was corrected by using a directed evolution approach that selected for enhanced photostability. This investigation yielded the mOrange2 derivative, which is about 30 percent more photostable than EGFP. mOrange2 also performs well as a fusion tag for cellular proteins, and the improved photostability should enable long-term imaging studies of cellular dynamics. The most useful red mFruit proteins, mCherry (610 nanometer emission peak) and mStrawberry (596 nanometer emission peak), have intrinsic brightness levels of approximately 50 and 75 percent that of EGFP, respectively. However, mCherry is more photostable than mStrawberry, so it is the preferred choice for cellular imaging. Recently, another red fluorescent protein, mApple, was generated in the same photostability screen that yielded mOrange2, and it features spectral characteristics close to mStrawberry, but with significantly improved photostability. The mApple variant is a rapidly maturing, bright, and photostable RFP that might prove to be better choice for tagging cellular proteins that are difficult to label (such as histones, tubulin, and connexins).
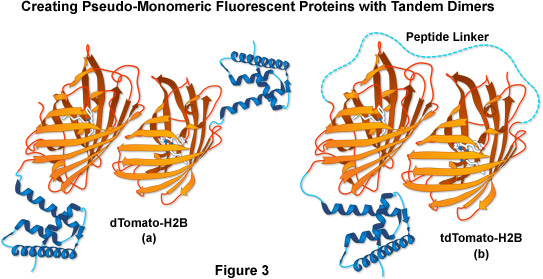
Another fluorescent protein that arose from the mFruit series screen was an obligate dimer variant named dTomato. To exploit its characteristics while simultaneously controlling the dimer formation, two dTomato units were joined (head-to-tail) through a sequence that encoded an optimized 12-amino acid linker, yielding a tandem dimer fluorescent protein, tdTomato (as illustrated in Figure 3). The tdTomato derivative is the brightest fluorescent protein yet reported, featuring an orange-red emission maximum at 581 nanometers, and is one of the most photostable fluorescent proteins under widefield illumination. Although the tandem dimer is twice the size of the other mFruits, it has proven useful in many fusions to cellular proteins, and is more easily detected, allowing it to be applied in live cell imaging studies at very low light levels.
The mFruit protein family was extended further by using a clever technique termed iterative somatic hypermutation. This approach yielded fluorescent proteins with the deepest-red emission of any of the mFruit proteins, which allow imaging in the far-red (630 to 700 nanometers) region of the visible light spectrum. The most useful probe is mPlum, which despite its low intrinsic brightness (approximately 10 percent of EGFP) has a good signal-to-noise ratio because of the reduced autofluorescence in this spectral region. The mPlum variant also has excellent photostability, and should be useful for multicolor applications when combined with fluorescent proteins emitting in the cyan, green, yellow, and orange spectral regions. These mFruit proteins, along with a multitude of new reef coral orange and red fluorescent proteins (discussed below) have filled essential gaps in the fluorescent protein color palette. The continued efforts to evolve proteins with optimal characteristics may ultimately yield that elusive RFP that has equivalent utility to EGFP. Among the important characteristics that still must be addressed are photostability, maturation time, brightness, acid resistance, and utility as fusion tags for cellular proteins that have been difficult to label. Although many new fluorescent proteins feature properties that meet or exceed those of EGFP in some of these categories, no single fluorescent protein yet discovered excels in all of them.
back to top ^Novel Fluorescent Proteins from Anthozoa
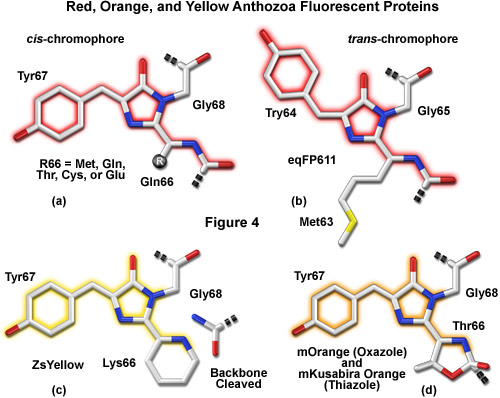
A variety of novel fluorescent proteins in virtually every color class have recently been cloned from Anthozoa and copepods, as well as a unique discovery of a family of GFPs in amphioxus. Researchers have extensively engineered these new probes to improve their utility for live-cell imaging. Essential to the understanding of spectral diversity in the wide range of fluorescent proteins discovered thus far are structural investigations of the stereochemical nature of the fluorophore and the effects of its surrounding environment on fluorescent properties. Unlike the Aequorea GFP derivatives, there appears to be a high degree of variation in the chromophores of red-shifted fluorescent proteins (Figure 4). Thus, even through the DsRed fluorophore configuration, termed planar cis (Figure 4(a)), appears to be the predominant structure in most proteins that emit in the orange and red regions, there are at least two additional motifs, planar trans and non-planar trans, which have been elucidated through x-ray diffraction studies.
A planar trans motif is found in the chromophore of the red fluorescent protein eqFP611, isolated from the sea anemone Entacmaea quadricolor (Figure 4(b)), which displays one of the largest Stokes shifts and red-shifted emission wavelength profiles of any naturally occurring Anthozoan fluorescent protein. In marked contrast, the non-planar trans conformation is characteristic of the non-fluorescent chromoprotein Rtms5 from Montipora efflorescens. In addition to the stereochemical variations, several of the proteins isolated from Anthozoa have markedly different chromophore structures from those of Aequorea GFP variants. The maturation of orange and red Anthozoa fluorescent proteins is believed to follow the same initial pathway as Aequorea GFP, but development continues with a second oxidation step that generates an acylimine moiety integrated into the peptide backbone between the amino acid residue preceding the chromophore and the first residue of the chromophore (usually methionine, glutamine, cysteine, or glutamic acid in Anthozoa fluorescent proteins). In several cases, the first residue of the chromophore also undergoes a cyclization reaction to form a third ring system (Figures 4(c) and 4(d)), which further influences the emission spectrum.
As further studies into the complex characteristics of fluorescent protein chromophores yield clues about the structure-function relationship with the polypeptide backbone, the task of genetically engineering more finely-tuned color variants and broadening the spectral range of useful proteins will undoubtedly become easier. Given that many of the amino acid triplets so far uncovered in chromophores from Anthozoa can give rise to huge variations in emission color (for example: MYG, 177 nanometers; QYG, 137 nanometers; TYG, 91 nanometers; CYG, 80 nanometers), it appears that there should be sufficient room in the fluorescent protein sequence space for additional mutations that will optimize color and many, if not all, of the other properties in fluorescent proteins derived from these organisms. The following discussion will target novel non-Aequorea fluorescent proteins that show potential in live cell imaging experiments.
back to top ^Blue Anthozoa Fluorescent Proteins
Similar to the situation with Aequorea fluorescent proteins, finding a blue variant with high brightness, pH resistance, and photostability from Anthozoa species has proven to be a challenging task. However, in the past couple of years several mutagenesis efforts have made significant progress in this area. Capitalizing on the fact that many Anthozoa red fluorescent proteins have chromophores that are formed via a blue intermediate, researchers applied random and site-directed mutagenesis on a variety of Anthozoa fluorescent proteins, including TagRFP, mCherry, and HcRed1, to convert these orange and red proteins into blue fluorescent proteins. The strategy was to introduce site-specific mutations that prevent maturation of the tyrosine-containing chromophore beyond the blue protonated intermediate, while simultaneously stabilizing the chromophore. The most promising candidate to emerge from this study was derived from TagRFP and named mTagBFP. When compared to blue Aequorea fluorescent proteins having histidine in the chromophore, mTagBFP demonstrates superior brightness, faster chromophore maturation, and higher pH stability. In terms of photostability, mTagBFP is less stable than EBFP2 under arc lamp illumination, but more than twice as stable when illuminated with laser light. mTagBFP has been demonstrated to perform well in most fusions with partner proteins and is an excellent FRET donor when coupled to green and yellow fluorescent proteins from both Aequorea and Anthozoa.
back to top ^Cyan Anthozoa Fluorescent Proteins
Several potentially useful cyan proteins have been isolated in Anthozoan species, and improved versions of these fluorescent proteins will likely see significant duty in live cell imaging. Derived from the reef coral Anemonia majano, the AmCyan1 fluorescent protein, which is now commercially available (Clontech), has been optimized with human codons for enhanced expression in mammalian cell systems. Originally named amFP486 (am for Anemonia majano; FP for fluorescent protein; 486 emission maximum) in accordance with a nomenclature scheme devised to simplify the classification of Anthozoan proteins, this variant exhibits a similar brightness level, but a significantly better resistance to photobleaching than Aequorea CFP. The absorption and emission maxima of AmCyan1 occur at 458 nanometers and 489 nanometers, respectively. Both peaks are shifted to longer wavelengths by 19 and 13 nanometers, respectively, compared to ECFP. Unfortunately, similar to most of the other reef coral proteins, AmCyan1 forms tetramers, which will significantly complicate attempts to employ this protein as a fusion tag or a FRET biosensor.
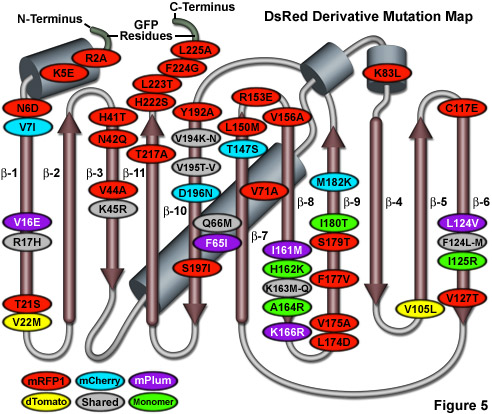
Illustrated in Figure 5 is a DsRed fluorescent protein mutation map showing alterations to the amino acid residues of useful variants superimposed on a topological layout of the peptide structure. The beta-sheets are numbered and depicted as thin, red cylinders with an arrow pointing towards the C-terminus, whereas alpha-helices are depicted by blue-gray barrels. Extensions of the N- and C- termini due to the addition of amino acids derived from EGFP to improve fusion performance are shaded in light green. Mutations are color-coded to represent the variants to which they apply: mRFP1 (red), mCherry (cyan), mPlum (violet), and dTomato (yellow). Monomerizing mutations for mRFP1 and the mFruit series of DsRed derivatives are shown in green while shared folding mutations are depicted in gray ellipses. Note that unlike the cluster of mutations surrounding the chromophore for Aequorea GFP variants, the DsRed mutations are distributed throughout the sequence. Unfortunately, due to the wide diversity of sequences found in Anthozoa fluorescent proteins, the DsRed mutation map outlined in Figure 5 does not correlate with fluorescent proteins from other reef corals and sea anemones.
Another potentially useful cyan Anthozoa protein, first isolated from an Acropara stony coral species, is a cyan-emitting derivative named Midoriishi-Cyan (abbreviated MiCy). MiCy was originally designed as the donor in a novel FRET combination with the monomeric Kusabira Orange fluorescent protein (discussed below) to generate a biosensor probe with high spectral overlap (Förster distance of 5.3). The fluorescence emission of MiCy features the longest absorption and emission wavelength profiles (472 and 495 nanometers, respectively) reported for any probe in the cyan spectral class. Furthermore, the high molar extinction coefficient and quantum yield exhibited by MiCy render the probe of equal brightness to Cerulean, although the spectra are far more sensitive to pH. Also similar to Cerulean, MiCy features a single exponential lifetime decay component with a time constant of 3.4 nanoseconds, which should be useful for measurements of FRET in combination with fluorescence lifetime imaging microscopy (FLIM). An unusual feature of MiCy is that it forms a homodimeric complex similar to the GFP variant isolated from the bioluminescent sea pansy, Renilla reniformis, rather than the obligate tetramer observed in most reef coral species. Although the dimerization motif may be a problem in some fusion proteins, it should be far easier (than a tetramer) to mutate MiCy into a true monomer.
Recently, a new monomeric cyan fluorescent protein having superior brightness, pH resistance, and photostability has been introduced for live-cell imaging applications of fusion partners, and as a FRET donor for yellow and orange acceptor fluorescent proteins in biosensors. Termed mTFP1 (monomeric teal fluorescent protein 1), the variant was produced from a synthetic gene library built around the tetrameric cyan protein, cFP484, originating from a Clavularia soft coral. Displaying red-shifted spectral profiles (excitation and emission maxima at 462 and 492 nanometers, respectively) when compared to most other cyan members of this spectral class, mTFP1 has a total of 31 amino acid substitutions relative to the wild-type tetramer. The red-shifted spectra were considered when classifying the new color as teal rather than cyan. Unlike members of the Aequorea cyan fluorescent protein group, which generally feature the aromatic amino acid tryptophan at position 66 in the chromophore, mTFP1 contains the classical tyrosine residue at this location. Substituting tyrosine for tryptophan reduces the broad fluorescence emission spectral width from approximately 60 nanometers to a more manageable 30 nanometers, a factor that is useful for reducing bleed-through in multicolor imaging experiments. mTFP1 has a high quantum yield and the relatively narrow emission spectrum strongly overlaps the excitation spectrum of the yellow and orange fluorescent proteins. The probe is also noteworthy because it fills the spectral gap between the cyan and green fluorescent proteins, and is optimally excited by the 457-nanometer argon-ion laser line that is available on most confocal microscopes. This new blue-green protein is also an excellent donor fluorophore for FRET studies using the Venus fluorescent protein.
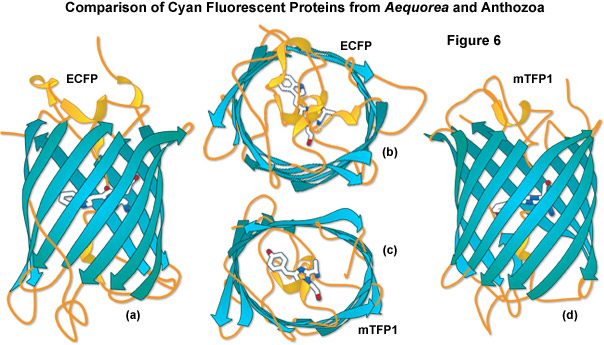
A comparison of the beta-barrel architecture between the high-performance Anthozoa and Aequorea GFP derivatives, enhanced cyan fluorescent protein (ECFP) and mTFP1, is illustrated in Figure 6. Although originating from vastly different sources, these proteins have many features in common. The chromophores are each positioned near the center of the barrel with the aromatic portion being oriented approximately perpendicular to the long axis of the molecule. As discussed above, the shape of the Anthozoa fluorescent protein viewed from the top or bottom is more ellipsoidal than that of the Aequorea variant. In addition, loops are more extensive and project farther away from the ends of the barrel for the jellyfish derivative. The chromophore of ECFP contains tryptamine in place of the wild-type tyrosine residue, whereas the mTFP1 chromophore retains the original tyrosine (along with its useful spectral characteristics) as discussed above. mTFP1 is much brighter and more photostable than ECFP, but it is also more problematic in some fusions where the Anthozoa variant demonstrates far less accuracy in localization.
back to top ^Green Anthozoa Fluorescent Proteins
A wide variety of fluorescent proteins emitting in the green spectral region have been isolated from Anthozoa, but others have also been found in copepods and amphioxus. Additional variants will probably ultimately be discovered in even more diverse organisms. One of the most promising of these probes, derived by random mutagenesis of a colorless protein isolated from Aequorea coerulescens, is known as aceGFP. The conversion of glutamic acid into glycine at position 222 (E222G) transformed the wild-type chromoprotein into a highly fluorescent species with a relatively symmetrical pair of spectral profiles having an absorption maximum at 480 nanometers and an emission peak at 505 nanometers. The high molar extinction coefficient and quantum yield of aceGFP combine to produce a brightness level similar to that displayed by EGFP. Demonstrated to exist as a monomer by electrophoresis and gel filtration, this protein is commercially available from several sources (Clontech and Evrogen, as AcGFP1 and AceGFP) with human-optimized codon replacements. Proper localization of fusion products targeted at specific subcellular components and organelles (such as filamentous actin, the Golgi, nucleus, and mitochondria) indicates that aceGFP is quite useful as a marker and could have potential for pairing with red-emitting proteins in a novel FRET combination. However, the photostability characteristics of aceGFP remain unknown, and there are no clear advantages to the use of this protein over the more common Aequorea EGFP and Emerald variants.
Several closely related GFP-like proteins have been isolated from an assortment of copepod aquatic crustacean species. The brightest of these probes, originally referred to as ppluGFP2, are commercially available (Evrogen) under the names CopGFP and TurboGFP (an enhanced variant). CopGFP is efficiently excited using an argon-ion laser or FITC filter set (absorption maximum at 482 nanometers) and produces green fluorescence at 502 nanometers with a brightness level approximately 30 percent higher than EGFP and much greater resistance to changes in pH. Reported to be a monomer in dilute solution, CopGFP matures significantly faster than EGFP and is ideal for applications as a fusion partner targeted at expression in subcellular regions of low pH. Limitations of this probe include the inability to isolate stable cell lines and the formation of aggregates in long-term cultures. An improved version, TurboGFP, derived from site-directed and random mutagenesis, retains the fast maturation kinetics of the parent protein with a slight loss in brightness and substantially lower resistance to acidic environments. Despite the improved folding kinetics and excellent optical properties of these proteins, however, photostability data have not been reported and no compelling evidence exists to demonstrate a significant benefit over the application of the extensively studied original GFP derivatives.
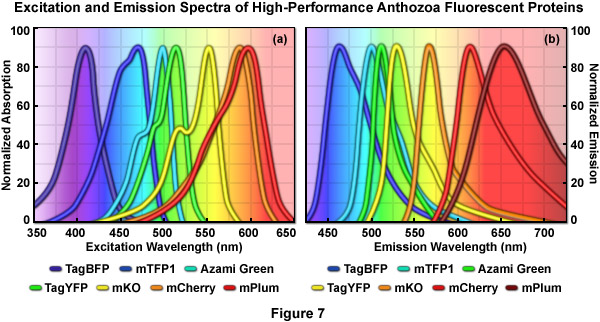
Normalized absorption (excitation) and emission spectral profiles for several of the more useful high-performance Anthozoan fluorescent proteins are presented in Figures 7(a) and 7(b), respectively. These variants, which are obtained from a diverse array of sources, cover a wavelength band approaching 200 nanometers, with emission maxima ranging from approximately 450 to 650 nanometers. Note the significant level of spectral overlap in the emission profiles (Figure 7(b)), which increases the complexity of segregating signals when more than two fluorescent proteins are imaged simultaneously. The absorption peaks of mCherry and mPlum feature a high degree of overlap even though the emission peaks are separated by approximately 40 nanometers (due to the unusually long Stokes shift for mPlum). One of the many engineering goals for creating advanced fluorescent proteins is to reduce the full width at half maximum (FWHM) of the spectral profiles to help ease the difficulty in separating closely overlapping signals during multicolor imaging experiments.
Surprisingly, a green fluorescent protein has recently been isolated from amphioxus, representing the first report of an endogenous fluorescent protein to be discovered in any representative of the deuterostome branch of the animal kingdom. Although not completely characterized, the amphioxus fluorescent protein (named AmphiGFP) features an amino acid sequence that predicts the standard beta-barrel structure and emits green fluorescence peaking at approximately 526 nanometers. Phylogenetic analysis indicates that AmphiGFP is more closely related to CopGFP than to the jellyfish versions. Several AmphiGFP variants were isolated from three species, which all express the protein in the anterior region of these shallow-water lancelets.
Green fluorescent proteins have also been extensively mined from reef corals and several of these are commercially available. A bright fluorescent protein termed Azami Green, bearing only a surprisingly scant (less than 6 percent) sequence homology to EGFP, was isolated from the stony coral Galaxeidae and has been demonstrated to mature rapidly during expression in mammalian cell lines. Likewise, one of the original Anthozoa reef coral proteins from Zoanthus has also been transformed into a commercial product (Clontech) under the trade name ZsGreen. The probes have absorption maxima at 492 and 496 nanometers and emission peaks at 505 and 506 nanometers, respectively, allowing visualization and imaging with standard lasers and filter combinations in confocal and widefield microscopy. However, similar to most of the other proteins isolated in corals, Azami Green and ZsGreen both exist as tetramers in the natural state, which significantly interferes with their use as fusion partners and as a FRET donor or acceptor in biosensors. To overcome the oligomerization problem, site-directed and random mutagenesis efforts were successful in creating a monomeric version of Azami Green (mAG), but this type of effort has not been reported for ZsGreen, although the protein has been re-engineered with human codons to optimize expression (resulting in a variant termed ZsGreen1). Because reliable fusion performance and photostability data is lacking, it is unclear whether either of these proteins will outperform EGFP in long-term imaging experiments.
Properties of Selected Anthozoa Fluorescent Protein Variants
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 1
A compilation of properties of the most useful Anthozoa-based fluorescent protein variants is presented in Table 1. Along with the common name and/or acronym for each fluorescent protein, the peak excitation (Ex) and emission (Em) wavelengths, molar extinction coefficient (EC), quantum yield (QY), relative brightness, photostability, and physiologically relevant quaternary structure are listed. The computed brightness values were derived from the product of the molar extinction coefficient and quantum yield, divided by the value for EGFP. This listing was created from scientific and commercial literature resources and is not intended to be comprehensive, but instead represents fluorescent protein derivatives that have received considerable attention in the literature and may prove valuable in research efforts. The excitation and emission peak values listed may vary in published reports due to the broad spectral profiles. In actual fluorescence microscopy investigations, the experimental brightness of a particular fluorescent protein may differ (in relative terms) from the brightness provided in this table. Among the many potential reasons for these differences are wavelength-dependent variations in the transmission or reflectance of microscope optics and the efficiency of the camera. Furthermore, the extent of fluorescent protein folding and maturation will depend on both the particular variant being used as well as the characteristics and localization of the fusion partner.
The sea pansy, an Anthozoa soft coral, is the source of several green fluorescent proteins that have been characterized in detail and are now commercially available. A protein isolated from Renilla reniformis that exhibits properties similar to EGFP is the best characterized of the probes in this class. Having absorption and emission maxima at 485 and 508 nanometers, respectively, in addition to a similar sensitivity to pH, the Renilla protein would be an excellent substitute for EGFP were it not for the fact that it is an obligate dimer. Aside from the oligomerization problem, Renilla GFPs may be useful in many applications and have been expressed in a wide variety of organisms, including bacteria, fungi, and mammalian cells. Versions with human codon sequences are available from the commercial distributors, as are derivatives optimized for expression in other species. There is a general lack of reliable data concerning extinction coefficients, quantum yields, and photostability for the commercial Renilla proteins, so valid comparisons to EGFP in terms of brightness and photobleaching are not possible.
In summary, although a wide variety of green fluorescent proteins have been obtained from species other than Aequorea, most exist natively as dimers or tetramers and have not been extensively characterized. Furthermore, it is unlikely that any Anthozoa green fluorescent proteins will ultimately be determined to outperform EGFP or Emerald in routine live cell imaging applications.
back to top ^Yellow Anthozoa Fluorescent Proteins
Unlike the situation for green, orange, and red fluorescent proteins, high performance fluorescent proteins having emission profiles in the yellow spectral region (ranging from 540 to 555 nanometers) are relatively scarce. The longest fluorescing Aequorea proteins have an emission maximum at 530 nanometers and appear green, rather than yellow, when viewed in a widefield fluorescence microscope. Although the potential for new discoveries of yellow and green fluorescent proteins in Hydrozoan species other than Aequorea is significant, only one candidate has surfaced so far. Isolated from the Phialidium jellyfish, a protein termed phiYFP is reported to demonstrate very bright yellow fluorescence (absorption and emission at 525 and 537 nanometers, respectively) and to be useful for N-terminal (but not C-terminal) fusion tags. An extraordinary feature of phiYFP is that the naturally occurring protein ironically contains two mutations that were discovered through protein engineering efforts on Aequorea fluorescent proteins. phiYFP contains the same mutation at position 64 (leucine) that was introduced during the creation of Venus (a high-performance Aequorea GFP derivative) to increase the folding efficiency, and also contains tyrosine at position 203, another site-directed modification of the native GFP that resulted in yellow fluorescence. This remarkable discovery of a natural similarity between the structure of phiYFP and genetically modified Aequorea proteins is a testament to the efficacy of protein engineering efforts directed at GFP to adjust the spectral properties. phiYFP has been optimized by random mutagenesis to produce a monomeric version without compromising the spectral properties, but this fluorescent protein has yet to see extensive use in live cell imaging or FRET applications.
Site-directed and random mutagenesis efforts on a monomeric Anthozoa fluorescent protein (mRFP1) from Discosoma have resulted in the creation of two monomeric reef coral derivatives with spectral properties falling in the range of Aequorea yellow fluorescent proteins, as described above. Named after similarly colored fruits, mHoneydew and mBanana both emit fluorescence in the yellow spectral region. The broad bimodal absorption band of mHoneydew (peaks at 487 and 504 nanometers) enables effective excitation with an argon-ion laser or standard FITC filter combination. However, the similarly broad bimodal emission spectrum (peaks at 537 and 562 nanometers) stretches into the near-infrared, hampering the use of this protein in multicolor experiments. In addition, a low extinction coefficient and quantum yield render mHoneydew the dimmest member of the monomeric yellow fluorescent protein cadre. The mBanana variant is only twice as bright as mHoneydew, but features much narrower excitation and emission spectra (peaks at 540 nanometers and 553 nanometers, respectively). Because both proteins exhibit relatively poor photostability, and mBanana is highly pH-sensitive, neither will probably find great utility in imaging experiments. Perhaps the most promising aspect of these probes is that the mere existence of mHoneydew (a cyan-type Y67W mutant) demonstrates that the tryptophan-based chromophore of CFP can undergo a further maturation into a longer-wavelength emitting species, which may serve as a basis for further improvement using site-specific mutagenesis or directed evolution.
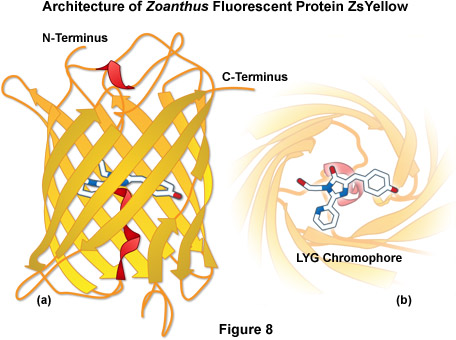
ZsYellow (originally referred to as zFP538) is a yellow fluorescent protein that was discovered in the Anthozoan button polyp Zoanthus. One of the most important features of the ZsYellow fluorescence emission spectrum is that the peak (538 nanometers) occurs almost midway between those of EGFP (508 nanometers) and DsRed (583 nanometers), presenting an opportunity to investigate proteins emitting fluorescence in the true yellow portion of the visible light spectrum. The ZsYellow chromophore features a novel three-ring system and peptide backbone cleavage due to the substitution of lysine for serine as the first amino acid residue in the chromophore tripeptide sequence (see Figures 4(c) and 8). As a result of the unique chromophore motif, the degree of conjugation observed in ZsYellow is intermediate between that observed with EGFP and DsRed (one double bond more than EGFP, and one less than DsRed), which accounts for the positioning of peak emission wavelengths in the yellow region. Unfortunately, ZsYellow exhibits a marked tendency to form tetramers when expressed in vivo, hampering the use of this protein as a fusion partner for localization investigations. Furthermore, the reduced brightness level of ZsYellow (25 percent of EGFP) also limits the utility of this reporter in fluorescence microscopy (the human codon-optimized version is commercially available from Clontech as ZsYellow1). The unique emission spectral profile of ZsYellow, however, should encourage the search for genetic modifications that alleviate the tendency to form tetramers while simultaneously increasing the quantum yield and extinction coefficient, an effort that could ultimately yield a high-performance monomeric yellow fluorescent protein.
back to top ^Orange Anthozoa Fluorescent Proteins
In the past several years, a relatively large number of potentially useful orange fluorescent proteins have emerged from various Anthozoa species. In one of the first examples, a protein named Kusabira Orange (KO) was isolated from the mushroom coral Fungia concinna (known in Japanese as Kusabira-Ishi). The sequence encoding KO was engineered to add ten amino acids to the N-terminus, resulting in a fluorescent protein having an absorption maximum at 548 nanometers (ideal for excitation with a 543-nanometer laser) while emitting bright orange fluorescence at 561 nanometers. In an effort similar to the strategy used to generate mRFP1, a monomeric version of Kusabira Orange (mKO) was created after site-directed and random mutagenesis of 20 amino acids. The monomer exhibits similar spectral properties to the tetramer and has a brightness value similar to EGFP, but is slightly more sensitive to acidic environments. However, the photostability of this fluorescent protein under arc lamp illumination is exceptional, making mKO an excellent choice for long-term imaging experiments. Furthermore, the emission spectral profile is sufficiently well separated from cyan fluorescent proteins to increase the FRET efficiency in biosensors incorporating mKO, and the probe is useful in multicolor investigations with a combination of cyan, green, yellow, and red fluorescent proteins. Recently, a fast-folding variant containing 8 additional mutations, named mKO2, was developed and should improve the utility of this probe for live cell imaging.
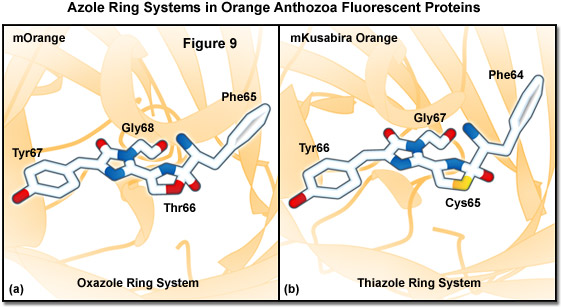
Similarities between the chromophores of mKusabira Orange and mOrange are illustrated in Figure 9. During protein maturation, both chromophores undergo a second oxidation step to produce an acylimine linkage in the polypeptide backbone (typical of red Anthozoa fluorescent proteins) followed by the spontaneous formation of a novel third ring system. In the case of mOrange, this third ring is an oxazole formed by the threonine residue at position 66, whereas in mKusabira Orange, the ring is a thiazole formed by the cysteine residue at position 65. Thus, these orange fluorescent proteins have emission profiles that are blue-shifted (from red to shorter orange wavelengths) relative to the red variants due to elimination of the conjugation between the acylimine carbonyl and the chromophore. This phenomenon is similar to the yellow emission arising from reduced conjugation observed in ZsYellow (discussed above) when the lysine residue at position 66 cyclizes with the acylimine to form a partially unsaturated piperidine ring system.
Another bright monomeric orange protein, named TagRFP, has been derived from the dimeric protein known as TurboRFP, which was originally cloned from the sea anemone Entacmaea quadricolor. TagRFP was generated by site-directed mutagenesis to replace several key amino acid residues involved in the dimer interface, while simultaneously performing random mutagenesis to rescue folding properties. Eight rounds of mutagenesis resulted in the final variant, which features excellent photophysical properties. TagRFP features an excitation peak at 555 nanometers, an emission peak at 584 nanometers, and is well tolerated as a fusion partner for many different proteins expressed in a variety of mammalian cell systems. The only drawback of TagRFP is relatively poor photostability under arc lamp and laser illumination when compared to other fluorescent proteins in this spectral class. However, during the same investigation that uncovered a highly photostable variant mOrange2, a similar mutagenesis strategy for TagRFP yielded a single mutation (S158T) that increases the photostability almost 10-fold. The resulting fluorescent protein, named TagRFP-T, has spectral properties similar to the parent and is among the most photostable of the fluorescent proteins yet discovered.
A novel orange fluorescent protein isolated from the Cerianthus tube anemone is commercially available (trade name cOFP; Stratagene) and has spectral properties that are similar to mOrange and mKusabira Orange, but like the other anemone proteins isolated to date, exists in solution as a tetramer. The brightness and photostability of cOFP have not been reported so this protein cannot be directly compared to other orange fluorescent proteins, and its utility will be further limited until it can be converted into a monomer.
back to top ^Red and Far-Red Anthozoa Fluorescent Proteins
The relentless search for a high performance red-emitting fluorescent protein to act as a complement to EGFP has begun to yield promising results. Red-emitting fluorescent proteins are particularly important due to the requirement for probes in this spectral region in multicolor imaging experiments as well as the fact that longer excitation wavelengths generate less phototoxicity and can probe deeper into biological tissue. Most of the fluorescent proteins in the red region of the visible spectrum can be imaged with the common TRITC and Texas Red widefield fluorescence filter sets, as well as cheap helium-neon (543, 561, and 594 nanometers) and diode (532 and 561-nanometer) lasers in confocal microscopy. After five years of unsuccessful mutagenesis efforts in the Aequorea GFP-derived proteins, the first real breakthrough occurred with the discovery of potentially fluorescent chromoproteins in non-bioluminescent Anthozoa coral species. To date, a wide spectrum of potentially useful red fluorescent proteins has been reported (spanning the emission wavelength range of 600 to 650 nanometers), many of which still suffer from some degree of the obligatory quaternary structure bestowed by their species of origin. Unlike the jellyfish proteins, most of the native and genetically engineered variants of reef coral proteins mature very efficiently at 37° C, presumably due to differing water temperatures of their respective habitats.
In addition to DsRed and its variants (discussed above), several additional red fluorescent proteins showing a considerable amount of promise have been isolated from the reef coral organisms. One of the first to be adapted for mammalian cell applications is HcRed, which was isolated from the anemone Heteractis crispa and is now commercially available as HcRed1 (Clontech). HcRed was originally derived from a non-fluorescent chromoprotein that absorbs orange light through site-directed and random mutagenesis. A total of six amino acid substitutions were necessary to create a red fluorescent species that matured rapidly and efficiently at 37° C (absorption and emission at 588 and 618 nanometers, respectively). However, similar to many other reef coral proteins, the resulting red fluorescent HcRed displays a tendency to form obligate tetramers when expressed in bacteria. Additional mutagenesis efforts resulted in a brighter dimeric variant, but a monomeric version of the protein has not yet been produced. In order to generate a derivative of HcRed that is useful in creating fusion vectors for localization studies, a tandem dimer of HcRed has been constructed. When fused to a gene product that itself forms biopolymers (such as actin or tubulin), the HcRed tandem dimer (tdHcRed) forms an intramolecular self-association complex (mimicking a monomeric tag) that apparently does not interfere with the biological activity of the resulting chimera. However, because the overall brightness and photostability of tdHcRed has not yet been improved, it remains a secondary choice for routine applications in live-cell microscopy.
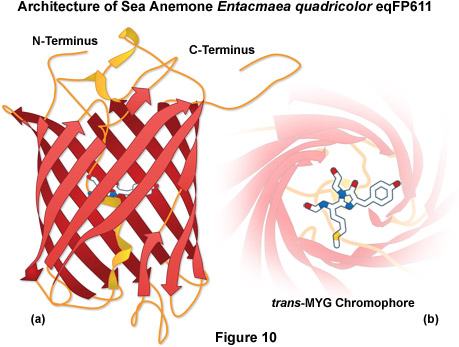
A potentially useful far-red fluorescent protein, termed eqFP611, was isolated from the sea anemone Entacmaea quadricolor and displays one of the largest Stokes shifts and red-shifted fluorescence emission wavelength profiles (excitation and emission maxima at 559 and 611 nanometers, respectively) of any naturally occurring Anthozoan fluorescent protein (see Figure 10). The quantum yield and extinction coefficient of eqFP611 combine to yield a probe approximately as bright as EGFP. During the in vivo fluorophore maturation process, which occurs in about 12 hours at 30° C (this protein does not mature at 37° C), the protein passes through a green intermediate state. After maturation, however, only a small fraction of this green species (less than 1 percent) can be detected. In contrast to other Anthozoan fluorescent proteins, eqFP611 has a reduced tendency to oligomerize at lower concentrations as evidenced by electrophoresis and single-molecule experiments, although at high concentrations the fluorescent protein does form tetramers. Site-directed mutagenesis efforts have yielded functional dimeric variants of eqFP611, but use of these fluorescent proteins is hampered by the low maturation temperature that is not compatible with mammalian cells.
Additional work on eqFP611 using random and site-directed mutagenesis has led to a series of red fluorescent proteins that feature long Stokes shifts and emission maxima extending to 639 nanometers. Introduction of several mutations into the parent fluorescent protein yielded a dimeric variant termed RFP611 (RFP for Red Fluorescent Protein), which was subsequently converted into a tandem dimer (tdRFP611). This improved version of eqFP611 features a high extinction coefficient and quantum yield that is significantly brighter than EGFP, but less photostable. In addition, a new far-red derivative (RFP639) and its tandem dimer were developed, but these fluorescent proteins are only about half as bright as EGFP. Continued efforts produced a bright, monomeric version of eqFP611, named mRuby, which contains 29 mutations relative to the parent. mRuby has excitation and emission maxima at 558 nanometers and 605 nanometers (similar to mCherry), respectively and is one of the brightest monomeric red fluorescent proteins yet developed. As an added bonus, the fluorescent protein retains the high Stokes shift of eqFP611 and is resistant to acidic environments (pKa of 4.4). Fusions of mRuby to over 30 targeting peptides and proteins have demonstrated excellent localization in most fusions.
Two additional reef coral red fluorescent proteins, AsRed2 and JRed, are commercially available (Clontech and Evrogen), but these probes form tetrameric and dimeric complexes, respectively, hampering their use in fusions. AsRed2 was originally isolated as a chromoprotein from Anemonia sulcata and modified through mutagenesis to yield a protein having an absorption maximum at 576 nanometers and an emission peak at 595 nanometers with a very modest quantum yield (0.05). Although the protein has been optimized with human codons for expression in mammalian cell lines, it exhibits only about 10 percent the brightness level of EGFP and the photostability has not been reported. The dimeric protein, JRed, was derived through extensive mutagenesis of a jellyfish chromoprotein to produce a novel red fluorescent marker with peak absorption and emission wavelengths of 584 and 610 nanometers, respectively. The probe has been demonstrated to produce useful fusion tags that tolerate dimers, but is unsuitable for expression in prokaryotes due to folding problems. JRed is about 25 percent as bright as EGFP and exhibits limited photostability when illuminated in the 560 to 580 nanometer region, but can be successfully employed for long-term imaging experiments when excited with a 543-nanometer laser.
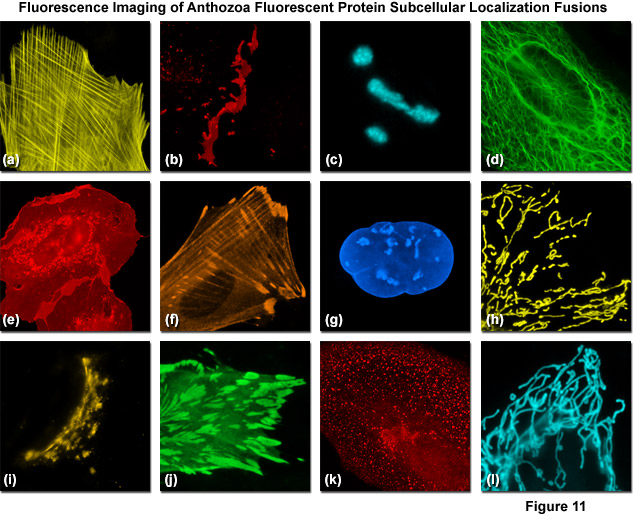
Presented in Figure 11 are fluorescence images captured in laser scanning confocal and spinning disk microscopy of several high-performance Anthozoa fluorescent proteins fused to subcellular localization targets. A fusion of mOrange2 to the actin-binding peptide domain known as Lifeact is shown highlighting the filamentous actin network in Figure 11(a). Figures 11(b) through 11(d) illustrate mApple, mTFP1, and mWasabi fluorescent proteins fused to connexin Cx43, fibrillarin, and cytokeratin, respectively. Figures 11(e) through 11(h) depict mRuby, TagRFP, TagBFP, and TagRFP-T fused to C-Src, zyxin, lamin B1, and a mitochondrial targeting signal, respectively. Finally, Figures 11(i) through 11(l) show mKO2, mWasabi, mCherry, and mTFP1 fused to a Golgi targeting peptide sequence, paxillin, clathrin light chain, and a mitochondria targeting sequence. In many cases the localization accuracy of Anthozoa-based fluorescent proteins is inferior to that exhibited by derivatives of Aequorea-GFP. This behavior is likely due to the fact that the monomeric Anthozoa proteins have all been derived from tetramers through extensive mutagenesis efforts, which often compromises other properties.
In a search for far-red fluorescent proteins, researchers applied a directed evolution and random mutagenesis approach to the Entacmaea quadricolor protein known as eqFP578 and selected for novel fluorescent proteins with deep red emission. This screen yielded a dimeric RFP called Katushka (emission maxima of 635 nanometers). Although only two-thirds as bright as EGFP, Katushka exhibits the highest brightness level of any fluorescent protein emitting in the deep red spectral window (650-800 nanometers), a region that is important for deep tissue imaging. A tandem dimer version of Katushka performs well in many fusions and is approximately 4 times brighter than mCherry and 20 times brighter than mPlum. Introduction of the four principal Katushka mutations into TagRFP generated a monomeric, far-red protein named mKate that has similar spectral characteristics (Table 1). The photostability of mKate is reported to be exceptional, and the protein displays brightness similar to mCherry, which makes it an excellent candidate for localization experiments in the far-red portion of the spectrum. Continued mutagenesis of mKate led to a brighter variant, mKate2, which is three times brighter, but retains similar spectral properties. mKate2 features excellent pH resistance and photostability, and transgenic studies in Xenopus embryos revealed that the protein has low toxicity for imaging applications in living animals.
Another far-red fluorescent protein, termed AQ143, has been derived from mutagenesis efforts on a chromoprotein isolated from the anemone Actinia equine. The excitation and emission maxima of AQ143 are 595 and 655 nanometers, respectively, and the brightness is comparable to mPlum. AQ143 features the longest emission wavelength maximum of any fluorescent protein yet reported, but it forms an obligate tetramer and thus, will probably see little action as a fusion tag. Regardless, the fact that fluorescent proteins are capable of emission at such long wavelengths provides hope that protein engineers will someday be able to fine-tune the emission properties of existing or as yet undiscovered fluorescent proteins into useful near-infrared probes.
Although promising imaging candidates are now available in every fluorescent protein spectral class from either Aequorea or Anthozoa species, there remains no equivalent to EGFP in terms of photostability, brightness, and performance in fusions, for much of the color palette. New additions to the blue and cyan region derived from several species exhibit substantially improved brightness and photostability, and many of the new orange proteins are excellent choices for long-term multicolor imaging. In addition, although brighter than EGFP, photostability is still suboptimal for all of the yellow proteins, whereas the red and far-red proteins are among the dimmest fluorescent proteins in all spectral classes. Furthermore, many of the supposedly “monomeric” fluorescent proteins do not, in some cases, localize as expected and are subject to aggregation artifacts. Even so, many of the newly reported fluorescent protein derivatives can be combined for dual and triple color imaging to yield excellent results. Given that most of the orange and red fluorescent proteins have only been introduced in the past couple of years, we remain optimistic that further studies into the complex characteristics of fluorescent protein chromophores will yield clues about the structure-function relationship with the polypeptide backbone to render the task of genetically engineering more finely-tuned color variants and broadening the spectral range of useful fluorescent proteins much easier.
back to top ^Increasing the Stokes Shift of Fluorescent Proteins
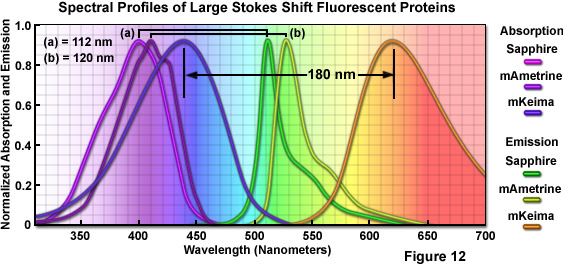
Imaging requirements for FRET, fluorescence cross-correlation spectroscopy (FCCS), and multicolor fluorescence have prompted protein engineers to target the separation distance between excitation and emission maxima (referred to as the Stokes shift) in fluorescent proteins through random and site-directed mutagenesis. In one of the first efforts, introducing the T203I mutation into wtGFP produces a variant, Sapphire, devoid of the minor excitation peak at 475 nanometers. Sapphire exhibits a dramatic Stokes shift of 112 nanometers, with excitation and emission maxima at 399 nanometers and 511 nanometers, respectively. A derivative with improved folding and brighter fluorescence, T-Sapphire (T for Turbo), was constructed by introducing four additional mutations. These variants should be excellent donors in FRET combinations with orange and red Anthozoa proteins due to their ability to be excited in the ultraviolet region. A more extensive mutagenesis effort on Aequorea blue fluorescent protein derivatives resulted in a new yellow variant, named mAmetrine, which exhibits a Stokes shift of 120 nanometers with excitation and emission maxima red-shifted by approximately 10 nm with respect to Sapphire (see Figure 12). mAmetrine was used to demonstrate the simultaneous application of two independent FRET biosensors in live cells. Both of the Aequorea-derived long Stokes shift variants should serve as the basis for mutagenesis efforts on related fluorescent proteins as well as those derived from other species.
Extending the Sapphire strategy to red fluorescent proteins, investigators used a far more rigorous approach to construct the longest Stokes shift fluorescent protein variant yet developed (180 nanometers) starting with a non-fluorescent chromoprotein derived from the Montipora stony coral. Mutagenesis of five residues surrounding the chromophore led to a red fluorescent protein having a bimodal excitation spectrum (peaks at 452 nanometers and 580 nanometers) with emission at 606 nanometers. An additional four mutations substantially reduced the 580 nanometers peak and blue-shifted the other absorption peak to 440 nanometers. This derivative, named Keima (after the Japanese chess piece), exhibits an emission maximum at 616 nanometers, but is tetrameric. Several more rounds of mutagenesis produced a dimer (dKeima) having similar spectral properties, and a monomer (mKeima; emission maximum at 620 nanometers) was subsequently obtained (Figure 12). mKeima exhibits limited brightness (similar to the value for mPlum) and requires a specialized filter combination for imaging, but it has been demonstrated to be useful in FCCS and multicolor imaging experiments. Recently, a tandem dimer version (tdKeima) has been created from the brightest variant, dKeima for use in fusions. Among the primary advantages of large Stokes shift fluorescent proteins is the potential for coupling several colors together, such as tdKeima, mCerulean, and EGFP, which can be excited with a single laser line in multicolor experiments, thus reducing the level of phototoxicity due to excitation illumination.
Conclusions
Although promising candidates are now available in every Anthozoa fluorescent protein spectral class (see Table 1), in most cases there remains no EGFP equivalent, in terms of photostability and other critical areas of performance (with the exception of pH stability). New additions to the blue and cyan region feature substantially improved brightness and photostability, and any of the orange fluorescent proteins are excellent choices for long-term multicolor imaging. In addition, although brighter than EGFP, photostability is still suboptimal for the yellow fluorescent proteins, whereas the red and far-red variants are among the dimmest in all spectral classes. Further compounding the problem is the potential for aggregation artifacts due to poorly folding proteins, regardless of the spectral class or supposedly monomeric characteristics. Even so, many of the fluorescent proteins listed in Table 1 can be combined for dual and triple color imaging to yield excellent results. Given that most of these proteins have only been introduced in the past couple of years, we remain optimistic that in the future, bright and photostable additions will become available for all spectral classes.
Contributing Authors
David W. Piston - Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee, 37232
Robert E. Campbell - Department of Chemistry, University of Alberta, Edmonton, Alberta, Canada, T6G2G2
Richard N. Day - Department of Cellular and Integrative Physiology, Indiana University School of Medicine, 635 Barnhill Dr., Indianapolis, Indiana, 46202.
Michael W. Davidson - National High Magnetic Field Laboratory, 1800 East Paul Dirac Dr., The Florida State University, Tallahassee, Florida, 32310.







